Healthy Soil with Graphite
Below are the third party findings, from Arizona State University, on the effects of soil amended with graphite from Greensmiths. Frankly, the results are staggering. However, we invite you to review them for yourself.
Greensmith’s Graphite Microbiome Analysis
C. Ryan Penton, Ph.D., Arizona State University
A. METHODS
A set of six pot replicates for a control (soil + plant) and treatment (soil + plant + graphite) experiment where the plant was butterhead lettuce (Lactuca sativa ‘Adriana’ ) was established. Lettuce was grown under artificial lighting in soils that originated from the Yuma, AZ cropping region. The soil was a sandy clay loam (58% sand, 20% silt, 22% clay) with a pH of 8.2, a cation exchange capacity of 10.4 meq/100g, total carbon of 0.23%, total nitrogen of 0.921%, porosity of 52.7%, organic matter content of 1.24%. Irrigation was performed with 200 mL of water as needed, typically once every six days with DI water supplemented with fertilizer. Briefly, 150 ppm N working fertilizer was prepared from commercial mix fertilizer (15N–5P2O5–20K2O Tap Hydro FeED; JR Peters, Inc., Allentown, PA). The pH was adjusted to 5.5 using sulfuric acid. Pots were arranged in a randomized block experiment. Following plant maturity, the plant and roots were removed and bulk soil was mixed in the pot. A subsample of 0.5 g of soil from each pot was used for DNA extraction using the Powersoil DNA extraction kit (Qiagen) followed by quantification on a Qubit fluorimeter.
For sequencing, targeted amplicon sequencing was performed on the Illumina MiSeq platform utilizing 2×250 chemistry. A total of 420,512 were initially obtained with 378,461 filtered and de-noised 16S rRNA gene sequences from 6 submitted samples with an average of 52,564 sequences per sample. Classification was based on the Greengenes database. Statistical analyses were performed based on Bray-Curtis dissimilarity matrices including a dummy variable after Hellinger transformation of raw sequencing data using PRIMER-E in order to test for significant differences between the control and graphite treatment.
The abundance of the soil microbial community was assessed utilizing 16S rRNA gene targeted primers 341F/797R with amplification performed using a Applied Biosystems Quantstudio 3 with SyBR Green chemistry. DNA extraction was performed using the DNEasy Soil DNA extraction kit and quantified on a Qubit fluorometer to enable conversion of gene abundances to per gram soil. qPCR was performed using E. coli K12 extracted plasmids containing the amplicon. All reactions were performed in triplicate. In order to assess the abundances of diazotrophs within the soils, we again implemented a quantitative PCR assay, this time targeting the nifH gene which encodes for a portion of the nitrogenase gene, essential for N fixation using the published and widely implemented Poly primers. All reactions were performed in triplicate and standards were based on cloned nifH genes for absolute abundances.
B. RESULTS
1. SEQUENCING
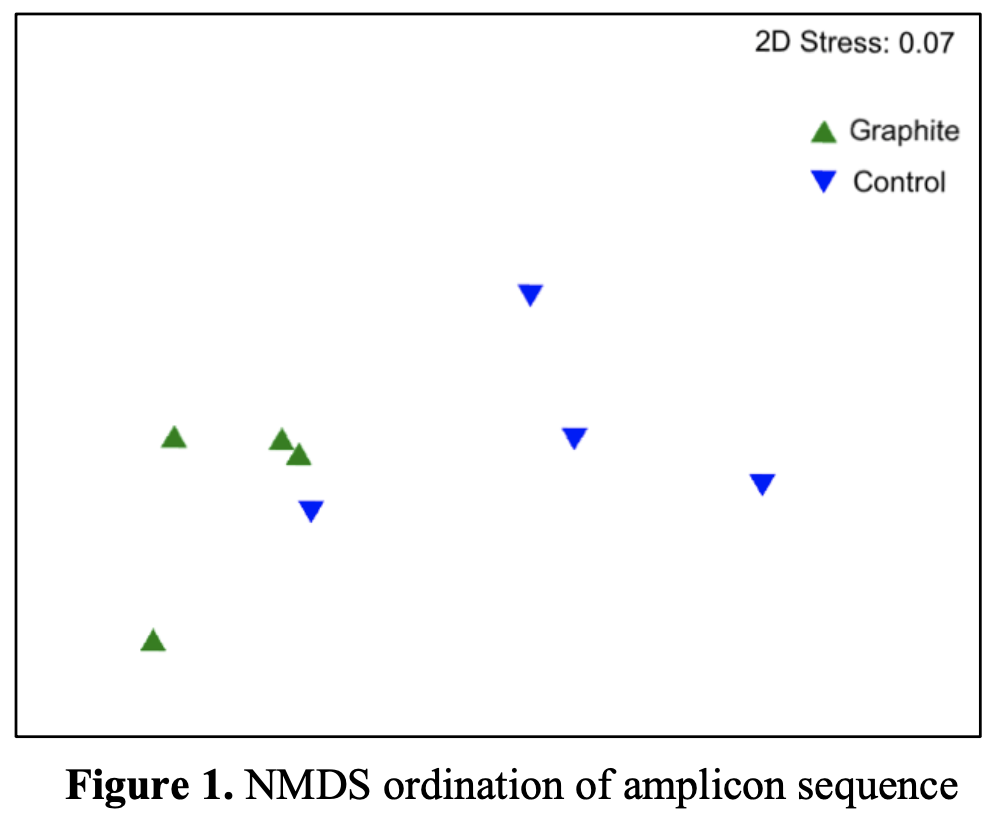
Permutational Manova (PERMANOVA) was used to test the significance of the treatment in regard to the composition of the overall bacterial community based on amplicon sequence variants (ASV’s) classified at the genus level. PERMANOVA revealed that the graphite treatment exerted a significant effect in altering the composition of the overall soil bacterial communities (Pseudo-F=5.97, p<0.01). Non-metric dimensional scaling (NMDS) ordination was used to visually compare the differences between samples and treatments (Figure 1). A clear separation was observed. Therefore, these data indicate that the application of graphite exerted a significant overall impact on the soil microbial community.
At the genus level, the top 40 ASV’s accounted for 42.6% of all sequences in the control and 71.2% in the graphite. This indicates that the graphite treatment homogenizes the bacterial community and favors the growth of certain bacterial lineages. Of these top 40 variants comparing the treatment and control., eight ASV’s were shared between both treatments, indicating that the differences between the control and treatment was principally due to recruitment of different bacterial lineages and not due to changes in abundances of bacteria that were already present.
The ‘core’ microbiome of the most abundant bacteria shared between the control and graphite treatment were: Bacillus (14.1%), Rubrobacter (2.1%), Microvirga (2.8%), Unknown Gemmatimonadaceae (1.8%), Vicinamibacteraceae (3.0%), Sphingomonas (3.3%) and Ammoniphilus (6.0%). In total, the ‘core’ microbiome of the ‘abundant’ bacteria constituted 11% of the control and 24% of the treatment, indicating that most shared taxa were bacteria of very low abundances. These most abundant bacteria from both treatments were further investigated to determine those that differed the most. Of these, the most remarkable influence of graphite was a strong recruitment of Bacilli (classified to different levels at varying confidences).
In total, there was a 7.3-fold increase in Bacilli within the graphite treatment, a 6.3-fold increase in Ammoniphilus and a 5-fold increase in Streptomyces. Conversely, graphite treatment resulted in a decrease in Bacteroidetes (1.50 to 0%), 5-fold decrease in unknown Nitrosomonas, and a bias against Gemmatimonadaceae (~2- fold decrease).
The largest group of bacteria that responded to the highest degree with graphite amendment were within the class Bacilli, a widely known plant growth promoting clade. Within the Bacilli, Paenibacillus prosopidis is capable of nitrogen fixation, and other Paenibacillus species are known to also exhibit biocontrol against broad phytopathogenic fungi, thus playing an important role in plant growth promotion. Multiple members of the Bacilli are also capable of nitrogen fixation, thus reducing fertilizer needs. Ammoniphilus were recruited by graphite and are aerobic obligate oxalotrophic bacteria which require oxalic acid as the sole organic C source as well as large quantities of ammonium ions, thus thriving in conditions where ammonium (NH4+) is in a free, unbound state in soil porewater. There was little change in Microvirga, another N-fixing bacterial genera.
Bacterial lineages selected against by the graphite treatment included the ~2 fold decrease in Gemmatimonadetes. These bacteria are known to be associated with plants and within the rhizosphere, have been found most associated with agricultural soils with some members have shown to be able to reduce N2O to N2 gas, thus playing a role in denitrification. Their abundances have been positively correlated with carbon, nitrogen and phosphorus availability in soils. Bacteroidetes were selected against, in contradiction with several previous lines of evidence that show a recruitment of these bacteria in the presence of graphene or graphite anodes in soil. Lastly, the reduction (5-fold) in Nitrosomonas reflects a potential decrease in ammonia oxidation (nitrification) in the presence of graphite. This would correlate with a previous study that showed decreased potentials for N loss through nitrification with the addition of various graphene nanomaterials.
2. QUANTITATIVE PCR:
a. ABUNDANCE OF THE TOTAL BACTERIAL COMMUNITY:
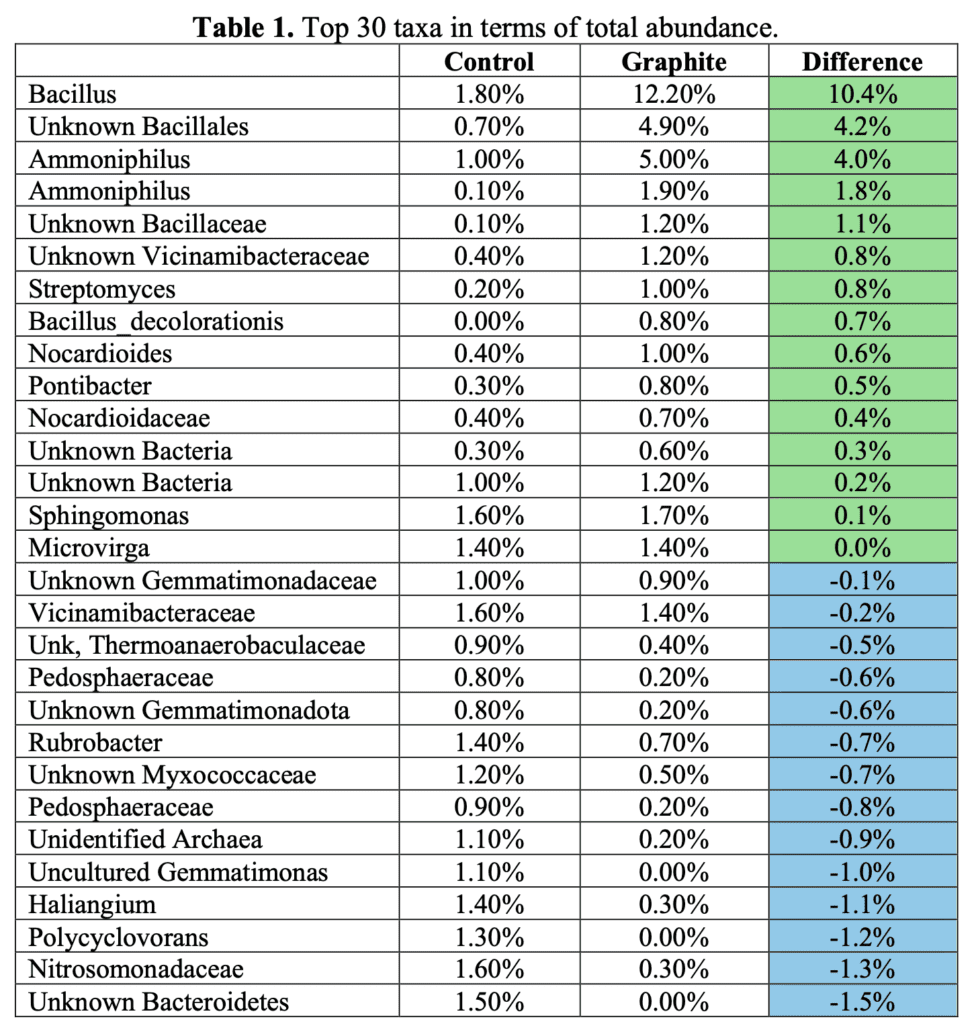
The overall abundance of the microbial community ranged from 6.58e5 to 2.05e6 copies per ng environmental DNA or 3.19e6 to 2.47e7 per g soil. In general, we most commonly use the copy number per g soil when we assess communities within soil. Nevertheless, both measurements reflected significantly higher (t-test, p<0.001 for ng DNA and p<0.001 for copies per gram soil) in the graphite treatment versus the control. This indicates that graphite addition is resulting in a larger microbial community over the 7-week pot experiment. We may have expected that if a larger community is present then it would be primarily those bacterial lineages which can utilize graphite as an electron acceptor, such as the Gemmatinomas, Burkholderia or Shewanella, for example. However, the sequencing results do not support this. In summary, a larger bacterial community was recruited with the addition of graphene. This higher microbial biomass is generally linked with a ‘healthier’ soil.
b. ABUNDANCE OF PUTATIVE N-FIXING BACTERIA:
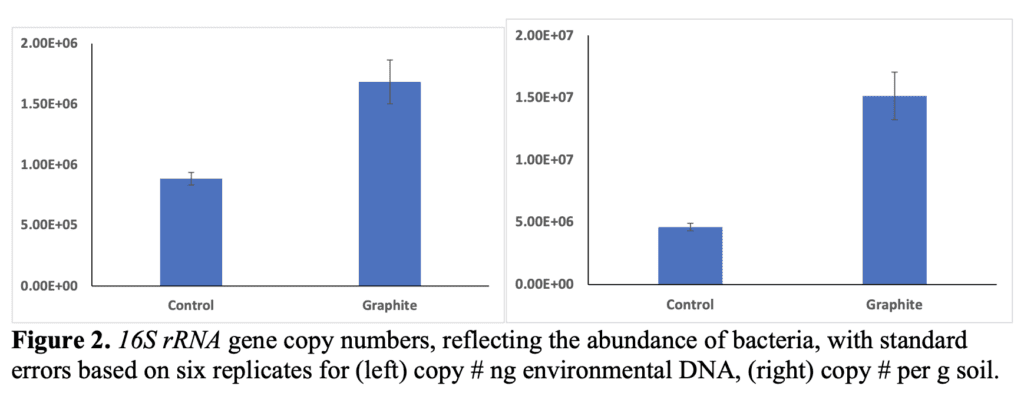
Abundances of putative N-fixing bacteria were estimated by quantitative PCR targeting the nifH gene. This gene encodes for part of the nitrogenase enzyme that is responsible for converting atmospheric N2 to NH3, which becomes NH4+ in soil solution. The fixed N is then available for uptake by soil microbes or plants. Abundances are of putative N-fixers since this assay is based on DNA and does not reflect whether the bacteria are alive or dead, simply that they possess the potential capability to perform this function.
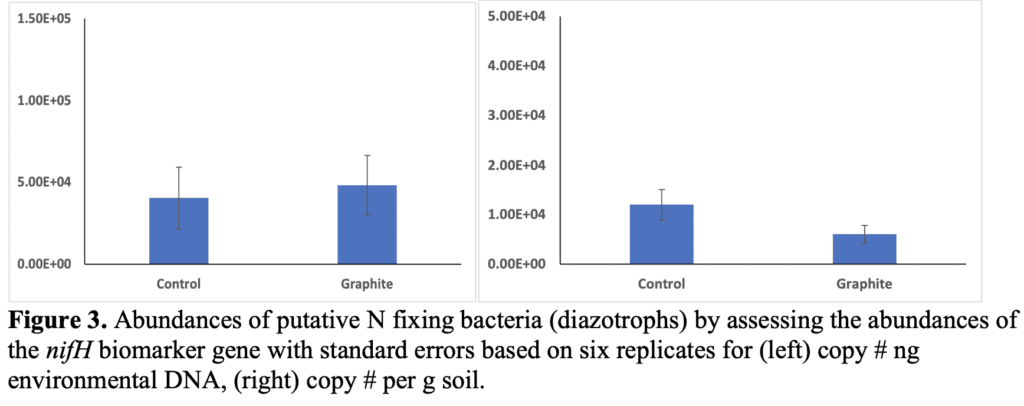
Results show that there were no significant changes in the abundances of N-fixing bacteria between the two treatments (t-test, p=0.8 (ng DNA) and p=0.12 (per g soil)). Thus, graphite addition did not significantly impact the N fixing bacterial populations. While there appears to be impact within the bulk soil, an impact may occur within the root rhizosphere, though that was not within the scope of this study. Lastly, qPCR was performed on extracted DNA targeting the ITS (inter-transcribed spacer) of fungal rRNA. In all cases but two the amount of fungi was below the limit of detection (~100 copies). We concentrated the DNA by 50% and were still unable to resolve quantifiable and comparable data from this assay. There have been times in the past where we have been unable to resolve fungi from these clayey desert soils so it is very likely that fungal abundances are very low in these soils which were taken from the 0-10 cm depth of a lettuce cropping field.
C. Conclusions
Together, these results indicate that graphite supplied by Greensmith’s has an appreciable impact on the soil microbial community. Application resulted in significant increases in total microbial community abundance with no significant impact on nitrogen-fixing bacteria populations. Community analysis revealed that graphite significantly altered the composition of the soil microbiome with a strong recruitment of Bacilli which harbor potential plant growth promoting rhizobacteria (PGPR). In addition, there was a recruitment of Streptomyces whose members are known plant growth promoters with some exhibiting biocontrol activities against pathogenic fungi. There was also a marked reduction in Nitrosomonas, a broad group of nitrifying bacteria, potentially pointing to decreased N losses through NO3- leaching. If this is the case, then this was also reflected by an increase in Ammoniphilus that thrive at higher NH4+ concentrations. Overall, there appears to be potential beneficial effects imparted on the soil with graphite addition caused by the recruitment of specific bacterial lineages.
END REPORT
Greensmiths Comments
In section: 2. QUANTITATIVE PCR: a. ABUNDANCE OF THE TOTAL BACTERIAL COMMUNITY:
The extent of the change is considerable and merits reiteration. Within just 7 weeks, the levels of beneficial microbial bacteria in the “control” samples—referring to the untreated soil—were significantly lower compared to those with “graphite treatment.” These findings clearly demonstrate that incorporating Greensmiths Graphite Sand (AMGS) will substantially boost the population of beneficial bacteria in the soil, resulting in a much healthier environment for plant growth. Below, you can observe the figures reflecting this rise in “good” bacteria.
3.19 e6 = 3 million 900 thousand
2.47e7 = 247 million 700 thousand
For more information on our Agri Minerals Graphite Sand (AMGS) and sales, contact us.

